RIT researchers use machine learning to better understand the pathways of disease
Cancer, Alzheimer’s, and other diseases follow a pathway in the human body. It starts at the molecular and cellular levels, and through a series of complex interactions can lead to the development and progression of disease.
At RIT, a new project funded by the National Institutes of Health (NIH) is using artificial intelligence to map the full journey of illnesses and discover entirely new disease pathways. If successful, the RIT research could transform how scientists understand disease and speed the discovery of new drugs and treatments for some of today’s most pressing health challenges.
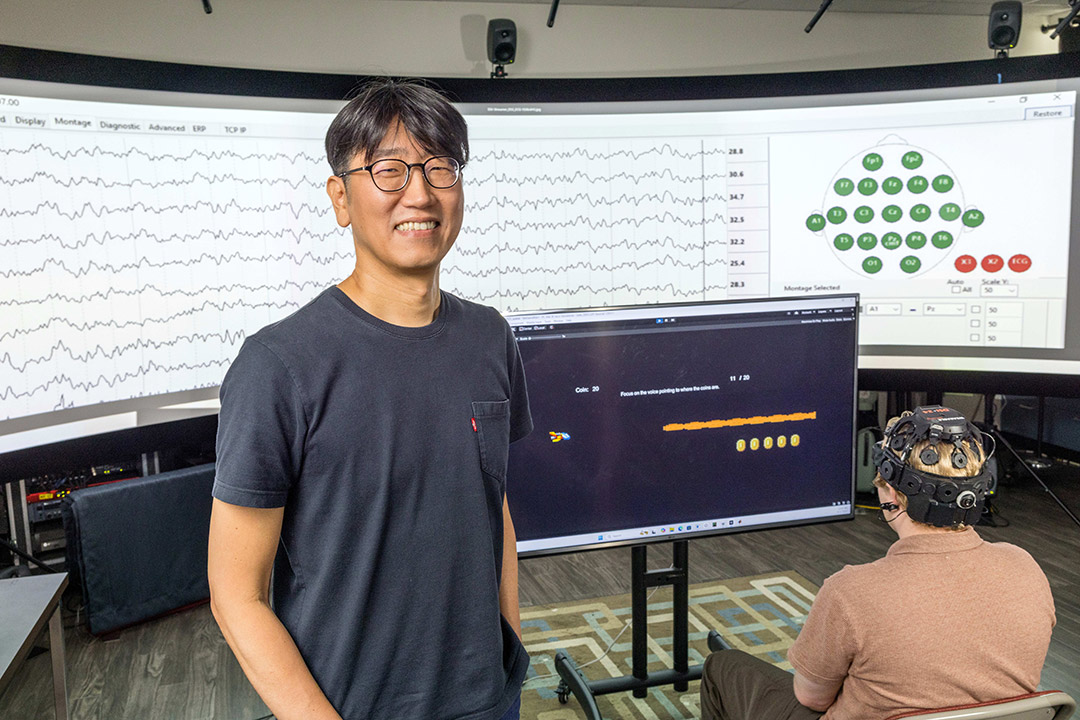
Rui Li, associate professor in RIT’s Golisano College of Computing and Information Sciences, received a nearly $1.8 million grant from the NIH for his project to advance research at the intersection of statistical machine learning and computational biology. Working with students, Li will design new machine learning models and techniques that enable disease pathway discoveries.
The challenge for Li is to create a holistic view of a complex molecular network of genes that are not well connected. If those genes have problems, it can have a cascade of effects. Atypical activities of these molecules, and their interactions, can contribute to disease.
“Using machine learning, we can identify new disease-associated molecules and biomarkers that could be potential therapeutic targets,” said Li. “This information is crucial for developing new drugs and could help caregivers.”
A second challenge is that diseases manifest differently in different body tissues, such as nervous tissue or muscle tissue. For example, disease-associated molecules that could collectively lead to one disease, like Parkinson’s, will manifest differently in different parts of the brain.
Li explained that current deep learning techniques are limited because they encode independently. Experts can make an average, but it will lose the uniqueness of the pathway making it hard to know how one molecule affects another.
“I propose a hierarchical model to capture the uniqueness at the local level, while also sharing complementary information at the global level,” said Li. “We will allow it to aggregate information from its neighboring nodes, which mimics what happens in the human body.”
Li’s recent NIH grant will support a new methodology developed to model molecular network data and its topological structure. For efficiency, these methods will integrate deep learning techniques with probabilistic inference.
The effort is powered by RIT’s Lab for Use-inspired Computational Intelligence, which Li directs. Four Ph.D. student researchers— Mahendra Singh Thapa, Paribesh Regmi, Sicy Ruochen Shi, and Jeevan Thapa—are currently helping analyze more than 100,000 molecular interactions.
“I’m fascinated by how perfectly our biological systems work, from tiny molecules to the whole organism,” said Thapa, a computing and information sciences Ph.D. student from Nepal. “I enjoy using machine learning to uncover hidden patterns and explore the mysteries of biology.”
The NIH-funded research will have two stages. First, the lab will conduct comprehensive studies of about 500 diseases, focusing initially on cancer, cardiovascular disease, and immune system disorders. Next, the work will expand to larger-scale applications across diverse disease types.
“I’m really excited to be working in this area and with Professor Li because it combines my passion for machine learning with important questions in computational biology,” said Regmi, a computing and information sciences Ph.D. student from Nepal. “I enjoy the challenge of developing algorithms that can not only advance AI research but also contribute to real-world impact in health and medicine.”
The five-year award comes through the NIH Maximizing Investigators’ Research Award program, which supports new and early-stage investigators developing innovative approaches to biomedical challenges. The project is called "Large-scale Disease Pathway Discovery by Integrating Tissue-specific Molecular Networks via Hierarchical Bayesian Inference on Graph Neural Networks".
Latest All News
- Exploring the complexities of life through a camera lensMeet the artist To celebrate their exhibition “of what might yet (have) be(en),” alum Lois Bielefeld will give an artist talk at 5:30 p.m. on Thursday, Oct. 9, in the University Gallery. A reception will be hosted from 4:30 to 7 p.m. on Oct. 9. See event details Lois Bielefeld ’02 (photographic illustration) finds inspiration in documenting domestic homelife, children’s tea parties, Sunday pasta lunches, and other personal moments. Capturing portraits from these unassuming scenes of everyday life comprises much of their portfolio. “I want to understand why we do what we do and what motivates us. Not just for big decisions or actions, but for the mundane and minutia of life,” said Bielefeld. “I think it's those minuscule daily decisions that create a person and fleshes out who we are.” Bielefeld, based in Milwaukee, Wis., is a senior photographer at Quad, a marketing experience company. After graduating from RIT, Bielefeld worked in New York City before moving back to Milwaukee in 2010. They have worked for commercial clients like Kohls, JCPenney, and Von Maur. In 2012, after solidifying their place in the world of commercial photography, Bielefeld began to lean back into personally meaningful projects. One of those projects, “of what might yet (have) be(en),” is on display in RIT’s University Gallery through Oct. 19. The exhibition is a culmination of three of Bielefeld’s interrelated bodies of work centered on gender identity: of what might yet (have) be(en); Androgyny; and I bring you a piece of horizon. Bielefeld aims to prompt exhibition viewers to think critically about their views of gender and consider the importance of supporting and protecting transgender and gender nonconforming individuals. “I believe that differences should be celebrated and embraced, but they are often feared,” they said. “One of my goals is that the person looking at my work will start reflecting on their own lives, and that it helps them understand other people’s experiences a little bit more.” Lois Bielefeld Lois Bielefeld stressed that she is not an expert in gender studies, but she wanted to listen and make space for people to share elements of their lived experience. Above is a portrait of Bielefeld from their House, Hold series from 2020. In 2013 and 2014, Bielefeld photographed 57 individuals for the series Androgyny. While working on the series, Bielefeld met Avery, a second-grade student who self-identified as a tomboy. Bielefeld was struck by Avery’s confidence and self-assuredness, and they decided to document his and another student’s collaboration with their teacher to create a gender inclusivity unit for their class. Creating that documentary video, titled Girl, Boy, Both, prompted Bielefeld to continue working with Avery for a new series, I bring you a piece of horizon, where Avery is photographed and interviewed every year on his birthday. This year marked Avery’s 19th birthday and the 12th portrait in the series. To create “of what might yet (have) be(en),” Bielefeld reconnected with the people they photographed for Androgyny, as well as five current RIT students, to take new portraits. The exhibition features a selection of portraits, short films, and immersive audio recordings, and it is the first time I bring you a piece of horizon has been shown in a public gallery. Exhibiting at their alma mater was a full-circle opportunity for Bielefeld because of the impact RIT had on their life. “My daughter was born in Rochester and when I became a mom, RIT supported me through that,” said Bielefeld. “It felt right to bring this work to campus.” Lois Bielefeld Two portraits included in the “of what might yet (have) be(en)” exhibition. On the left is a portrait of Cam taken in 2025. On the right is a portrait of Valentine taken in 2025. RIT Photo House, supportive and passionate faculty members like Denis Defibaugh and Allen Vogel, and the “immense and immersive” photography culture also contributed to Bielefeld’s fond memories of RIT. Bielefeld said many highlights of their career stemmed from their personal artistic practice. In 2015, Bielefeld was named the first winner of Museum of Wisconsin Art’s Artist-in-Residence Program and spent three months living and creating art in Luxembourg. In 2023, the Los Angeles County Museum of Art (LACMA) accessioned 10 photographs and three video works by Bielefeld into its permanent collection. The biggest “mega-moment” of their career, Bielefeld said, involved a deeply personal series titled To commit to memory. The work focused on their parents’ relationship with their home and everyday life and explored how religion added a “simmering intensity” and “chasm of ideological difference” to their family dynamic. Thank you Jesus, for what you are going to do, a video from the series, was selected for The National Portrait Gallery’s triennial Outwin Boochever Portrait Competition in 2022. The “mega-moment,” Bielefeld said, was having their mom and daughter watch the video work together in the National Portrait Gallery. To see more of Bielefeld’s work, go to their portfolio website. Go to the University Gallery website and opening reception event page for more information about the exhibition and artist talk on Oct. 9.
- RIT Dubai, SmartKable partnering to advance smart grid technologiesRIT Dubai is partnering with SmartKable Powerline Solutions, a New York-based company, and its Middle East and Africa arm SmartKable Equinova, to develop an AI solution that will enable a smarter electric grid. As part of the agreement, RIT Dubai takes on a small equity stake in SmartKable Equinova to allow for a long-term, beneficial collaboration for all parties involved. Faculty and students will collaborate with the company to advance smart grid technologies by integrating generative AI and predictive analytics into SmartKable’s Line Analyzer software. Students are heavily involved, with some doing capstone projects through Dubai’s Smart Energy Lab, and a postdoctoral student in Rochester who is providing architectural oversight and integration. Jinane Mounsef, chair of the electrical engineering and computing science department at RIT Dubai, who is one of the project’s leads, said the group is aiming to develop solutions that automate power assessment reporting with AI-driven insights, provide real-time smart notifications for grid reliability, enhance predictive capabilities to forecast failures and optimize planning, and support intelligent demand-supply control for resilient energy systems. “This initiative highlights how, together, academia and industry can transform challenges into opportunities for innovation and sustainable growth,” said Mounsef. SmartKable’s Line Ranger network measures line voltage, current, and related parameters on lines in real time. It can identify, track, and trace losses, separate commercial from technical issues, enable dynamic line rating, and predict the remaining lifetime of power lines. “We want to help the grid successfully evolve in spite of all the new electrification and items that are happening,” said Bill Collins, co-founder of SmartKable Powerline Solutions. “We ultimately want to turn the grid into something that is intelligently controlled by AI.” Collins explained that at the end of the project, the team expects that they will be able to start utilizing the solution commercially with customers. “The need for a smart, efficient, and predictable grid is a universal, global issue,” said Wojtek Bulatowicz, managing director of SmartKable Equinova. “This is why SmartKable now has its operations in the United Arab Emirates to serve the Middle East and Africa markets.” SmartKable is based in Skaneateles, N.Y., and already has deployed its Line Ranger devices around the world. The company has previously connected with RIT through the co-op program, and it was interested in partnering with Dubai after learning about the campus’s experts in engineering and AI. The agreement between the campus and the company is key to providing RIT students with the experience of real-life scenarios and the support to find solutions that can be used worldwide. For more information on the Dubai campus, go to the RIT Dubai website. More information on the Line Ranger Network can be found on the SmartKable Powerline Solutions website.
- NTID researchers work to incorporate accessible AED machines on campusIn emergencies, Automated External Defibrillators (AEDs) help restore life until first responders arrive by giving users auditory prompts to operate the machines. But audio prompts and descriptions are virtually useless for deaf and hard-of-hearing individuals who may need to use AEDs to help save a life. Researchers from RIT’s National Technical Institute for the Deaf have identified an AED that includes visual and text prompts, making it accessible for deaf and hard-of-hearing users and those not familiar with the English language. As a result, accessible AED machines are now available at four campus locations: the first floor of Lyndon Baines Johnson Hall, Student Development Center, Hugh L. Carey Hall, and in a student residence—Ellingson Hall Tower A. Remaining units throughout campus will be replaced with accessible AEDs at end of life. “It’s terrific that AED machines have become widely available, including at RIT, but I’m deaf and what if I was alone and needed to use it on someone?” said Elizabeth Ayers, senior lecturer in NTID’s Department of Science and Mathematics and a licensed clinical sonographer. “I can’t hear the auditory descriptors such as the beeps and other background sounds, and neither can the hundreds of other deaf and hard-of-hearing people on our campus. We knew that something needed to be done in order to make these lifesaving machines user-friendly for just about anyone here.” Ayers and Wendy Dannels, research associate professor; Tiffany Panko, executive director of NTID’s Deaf Hub; and alumnus Menna Nicola ’25 (human-centered computing) worked as a team to investigate FDA-approved devices that portrayed the most visually accurate instructions and imagery. Thirty-two devices from seven manufacturers were examined. Of these, three models were identified as meeting accessibility criteria. RIT selected one for replacement and deployment across campus, with the guidance of Christopher Knigga, director of Facilities Services and Sustainability at NTID, and Jody Nolan, RIT’s manager of Environmental Health and Safety. Nicola, an associate IT analyst at Dow Chemical, was a student when the research began. “I didn’t really know much about AEDs before this project began, but I was thrilled at the opportunity to conduct research that directs impacts me and other deaf people,” she said. “The most enjoyable part of the project for me was the result that led to finding a great match for an accessible AED for deaf and hard-of-hearing individuals.” According to Ayers, next steps include reaching out to other schools, agencies, and organizations focused on deaf and hard-of-hearing individuals to share their research on the accessible AEDs. “RIT was on board with installing these accessible AEDs based on our work,” said Ayers. “It’s our team’s responsibility to be sure that other organizations know the options that are available to them. And hopefully, to take this one step further, manufacturers will continue to work alongside deaf researchers to develop and continually innovate other healthcare products that are fully accessible.”
- ‘Reporter’ builds momentum as a platform for student voicesRIT’s student-run Reporter Magazine is working to returning to its roots as a primary information source by and for the RIT community. As it heads into its 75th year in 2026, Reporter has added frequent news updates online between its monthly print publication, has an editorial staff interested in communications and journalism, is seeing an interest in students wanting to join the staff, and is again winning awards for its work. Mariella Santiago, a fourth-year journalism major from Pittsford, N.Y., was elected editor-in-chief in January. “I was interested in making Reporter more of a journalism publication,” she said. “We have a more streamlined process of how we do our print publications, with cycles every month. We make sure we are published on time and people meet their deadlines. I’m really proud of this team with what they’ve done.” About 50 students with a variety of majors are on the staff and get paid for their work. Some are freelance writers or photographers, and some receive a stipend. “We’re always looking for more people to help out with our publication,” she said. The organization is editorially independent and student-run, with an advisory board that offers suggestions as needed. Some revenue is collected from advertising, but the bulk of funding is provided from RIT. All of the editorial content comes from the staff. The staff meets weekly for a newsroom brainstorming session at 5 p.m. each Thursday in its office, room A-730 in the basement of Campus Center. They hold organizational meetings at 4 p.m. Tuesdays. Any RIT student or employee is welcome to attend. “We talk about what’s happening nationally and globally, and try to relate those issues to RIT, and what’s trending on campus,” Santiago said. “We want to view issues from a student’s perspective.” There are 3,000 print copies distributed free each month around campus, and that content is also put online and on social media. Staff members have experimented with videos and podcasts and hope to provide photography and videography workshops to students. Tom Dooley, former program director for RIT’s journalism option, has been Reporter’s faculty adviser for three years. He said this staff has made “an intentional effort to really serve the student body. They take a serious look at what the people in this RIT community want to know about, and how they can deliver that to them.” Dooley said the staff has hired more visual journalists, more social media managers, rebuilt their website on a platform with more visual capabilities, and increased their presence on Instagram. “I think they are doing good work now and are taking the job of being a voice for the students seriously,” said Dooley, a former photojournalist, television producer, and filmmaker. “They know their role is important here at RIT to serve the student body through a perspective they can really connect with in an era of local news deserts, with fewer and fewer journalists covering local communities. Reporter is here to add value to students’ lives and occasionally check the administration. And that’s OK for that to happen.” Dooley is quick to say he’s not the staff’s boss and has no editorial say or prior review of what gets published, but he does sit in their planning meetings and supports them if they have any challenges or can help them in the creative process if they ask for guidance. One suggestion they followed was to create a manual for the staff with clear editorial guidelines and editorial ethics. Dooley has also traveled with several Reporter staff members to conferences by the Associated Collegiate Press in Minneapolis and College Media Association in New York City in the past three years to learn about best practices among other college publications. Reporter has won awards at the conferences. A 12-member advisory board, which includes faculty and administration members, communications professionals, and alumni who work in media, also meets with staff members periodically, providing guidance and feedback to the students. Dooley said he’s encouraged that some staff members are starting to collaborate with local professional organizations, getting co-ops as they learn journalism first-hand and even getting bylines. Liam Conroe, a journalism major from Jamestown, N.Y., serves as Reporter’s copy managing editor. He completed a co-op as a reporter for Rochester Beacon and hopes to possibly become a sportswriter after college. Noah Gallo, a second-year cybersecurity major from Danbury, Conn., joined the staff as a writer in March after a friend and Dooley, who taught a communication course he was in, suggested he join. He’s written two stories and two reviews since then. “While he doesn’t have a vision to become a career journalist, he says he can take aspects of what he’s learning at Reporter and apply them to his major. For the October edition, he’s writing a review of the videogame Cyberpunk 2077. “I never thought I would get to write an article about my favorite game,” Gallo said. “Just the fact I can and that people will read it is so cool to me.” Santiago, who completed an internship with News 10 (WHEC-TV) in Rochester, is gratified at the staff’s work and the product they are producing each month. “When I see it in print and see my editor’s note and my signature there, that is all worth all the work I put into it,” she said. “Seeing the final product and seeing people reading it around campus is always the most rewarding thing.”
- Audio engineer awarded funding to improve and personalize hearing devices and headphonesHwan Shim is using technology to personalize audio experiences because human ears are all different. Shim, an assistant professor of electrical and computer engineering technology, is developing new audio technology that could decrease extraneous sounds in noisy environments. This could improve applications from arts performances to personal devices such as hearing aids. “We often make technology and equipment with ‘average’ ears in mind,” said Shim. “But what if we can personalize this technology when we wear certain headphones or other devices? What if we can add personal effects that can still be natural and more spatial? At RIT, we are pushing this field.” Shim was recently awarded National Institutes of Health funding of nearly $750, 000 for “Semantic-based auditory attention decoder using large language models,” a project to determine how individuals distinguish sounds, and how the brain helps ‘censor’ sound that is non-essential to individuals, including those who need devices to improve hearing. Addressing sensorineural hearing loss requires more than amplification, and some auditory devices such as cochlear implants, while advanced, continue to improve by differentiating speech from background noise, Shim explained. His project, which began in August, could advance this speech-in-noise perception by mimicking the brain’s ability to discern and decode auditory messaging and sounds. His research team will use machine learning and integrate large language models to determine EEG-based selective attention decoding to improve hearing devices. Traci Westcott/RIT Biomedical engineering undergraduate, Danny Teets, part of the audio engineering research team, wears a prototype EEG device to explore how the brain censors sound in noisy environments. Today’s audio devices are sophisticated, but some underperform in noisy or distracting environments. To address these challenges, Shim and his research group in the Music and Audio Cognition Laboratory, based in RIT’s College of Engineering Technology, developed a prototype headset to provide neurofeedback information. The device is being used to record signals indicating how the brain recognizes needed information and separates out unwanted distractions. Shim’s work leverages the acoustic enhancement technology called Active Field Control (AFC), an immersive sound technology developed by Yamaha Corp. and RIT engineering colleague Sungyoung Kim, which is used in live performance spaces, such as auditoriums, and in virtual spaces, including virtual reality and gaming systems. Shim’s early doctoral research focused on spatial impressions, including reflections and reverberation that underpin such technologies. Coupled with AFC technology, he continues to explore spatial audio in various contexts, and these systems allow his research team to recreate realistic, noisy environments to develop, and test individualized selective attention decoding techniques. “The state of the industry is in immersive audio,” said Shim, who is an expert in applied acoustics, auditory cognition, and neuroscience, and who has industry experience as an engineer at Samsung. "Using field control processors, we can actively change the sound space, for example, during a live performance. We are also seeing changes in listening habits, and we are making better technology for immersive audio to give a more natural feeling of sound.” Shim said that although today’s systems are maturing, there are still gaps and limitations to presenting realistic, although enhanced, sound. Over the course of the project, the team will work to further three specific areas: improvements to their prototype device; expansion of the training program used to distinguish sounds; and refinements to analytical methods once data from the system are captured. Shim will also be exploring how individuals contextualize information, including sound, and how audio systems then incorporate these distinctions. This is an engineering challenge Shim sees as a natural evolution of audio technologies that have progressed from early recording devices to those with the most sophisticated noise-reducing functions. “We know that people have their own experiences and intimacy with sound. They can perceive differences,” he said. “We also know that the technology can decode the target speech from noise. Our idea is to clarify meaning of language to be more context-based. We are still figuring out what is going on in the brain. This can be next generation technology.”
- RIT faculty member and students create peace art installation at Woodstock siteThe rolling Bethel Woods hills that once hosted the iconic 1969 Woodstock music festival now holds a new architectural tribute to peace. The Peace Pavilion, designed and built with the leadership of RIT architecture Professor Amanda Reis, invites visitors to pause, reflect, and immerse themselves in the legacy of a site that continues to symbolize cultural change. Reis, in her second year as a faculty member in the Golisano Institute for Sustainability’s Department of Architecture, partnered with longtime collaborator Eduardo Aquino of the University of Manitoba to submit a proposal to BuildFest 2: Peace Rises, a part of the Bethel Woods Art & Architecture Festival. The event, held Sept. 10-14, called on university professors to create “peace structures” of varying scales. Their submission, entered under their research and design practice AREA, was selected for the largest commission. “It was a real honor to be selected,” Reis said. “We saw it as an opportunity to combine design innovation with sustainability and to involve students in a transformative learning experience.” Set against the backdrop of Filippini Pond and an adjoining forest, the pavilion occupies an Instagram-worthy location on the Bethel Woods grounds. The pavilion’s form was inspired by the yin-yang symbol, a representation of interconnected opposites and balance. Reis and Aquino reinterpreted it into a cubic structure that offers a cocoon-like feel, while also offering spectacular views. Youngjin Yi RIT architecture students gained hands-on experience assembling prefabricated panels for the Peace Pavilion. Those visiting the structure will step into a light-filled space whose walls are inscribed with phrases curated from poetry, literature, and music. Some lyrics also come directly from Woodstock-era artists, a connection to the area’s past. Western Red Cedar was chosen as the primary material because of its beauty and resilience. The material strengthens over time, meaning the Peace Pavilion could endure for years. “Many of the BuildFest structures are temporary, but we wanted to increase the longevity of the Peace Pavilion,” Reis said. “The material gives it that possibility.” Reis emphasized that the project’s success was the result of many minds. While Reis and Aquino established the design, the project became a hands-on experience for RIT architecture students. The project served as an early design-build exercise for Reis’s Architecture Studio course, a master’s architectural design class that typically emphasizes theoretical projects. The Peace Pavilion offered the students a rare chance to see ideas translated into reality. The student team included Noah Baldon, Maddy Bortle, Anna-Leigha Clarke, Ryan Denberg, Sydney Fox, Gabriela Hernandez, Gil Merod, Mackendra Nobes, Julia Resnick, and Youngjin Yi. “The collaboration exposed students to the full cycle of design-build, from concept to construction,” Aquino said. “They encountered unexpected challenges and learned how to improvise solutions during installation, which is where the deepest learning often happens.” In addition to the student team, staff from the SHED—Michael Buffalin, Jim Heaney, and Chris Vorndran—played a key role in fabricating the engraved cedar boards, before it made the four-hour trek to the Catskills. Ralph Gutierrez, an architectural intern, also contributed to the design development, while Yi captured photography throughout the process. “All of the dimensional wood was fabricated at 10-foot lengths to minimize waste, and the design required tight tolerances that pushed us to be meticulous during assembly,” Aquino said. “Those details emphasized the balance between sustainability and craftsmanship.” Youngjin Yi Inside the Peace Pavilion, engraved phrases line the cedar walls, which include lyrics from popular Woodstock performers. During the build, Reis mentioned that a couple who had attended Woodstock stopped by in their original festival T-shirts, eager to step inside the new structure. The pavilion offers potential for several outdoor activities. “I can imagine people reading, meditating, or just waiting for friends to finish kayaking or swimming nearby,” Reis said. “As architects, we can anticipate some uses, but the most exciting thing is what we can’t predict.” Fox, an architecture graduate student from southern Maine, said the entire process taught her important lessons. “I see architecture as a tool that can bring people together, communicate ideas and stories, and challenge the status quo,” said Fox. “Building this Peace Pavilion with others helped me see the power of collective effort and how creating something together can forge meaningful bonds between people, giving a project even more significance beyond its final result.”